 Recognition of MicroRNA Precursors and Targets
Recognition of MicroRNA Precursors and Targets
Participants: Wynne Hsu, Coral Lai, Mong Li Lee, Limsoon Wong,
Lianghuai Yang, Yun Zheng, Stanley Ng
Background
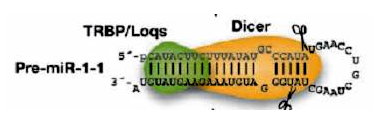
MicroRNAs (miRNAs) are small non-coding RNAs of about 22 nucleotides long.
MicroRNA transcripts, which may be produced by RNA polymerase II or III,
often fold to form stem loop structures, and become what are called
primary miRNAs, or pri-miRNAs. In the nucleus, the Drosha RNase III
endonuclease cleavages both strands of the stem at the base of the
primary stem loop, and produce the pre-miRNAs. Then, in cytoplasm,
a second RNase III endonuclease, Dicer, together with its dsRNA-binding
partner protein makes a second pair of cuts and defines the other end of
the mature miRNAs, which produces the miRNA:miRNA* duplex. Finally,
the miRNA stand is separated from the duplex by the helicase and forms
the mature miRNA molecules. The mature miRNAs are then loaded to RNA-induced
silencing complex (RISC), which binds the 3' untranslated region of
messenger RNAs of the miRNA target genes to repress the production of
related proteins.
MicroRNAs play an important regulatory functions in eukaryotic gene
expression through mRNA degradation or translation inhibition.
The regulatory functions of miRNAs range from cell proliferation,
fat metabolism, neuronal patterning in nematodes, neurological diseases,
modulation of hematopoietic lineage differentiation in mammals, development,
cell death, cancer, and control of leaf and flower development in plants.
An miRNA downregulates the translation of target mRNAs through base-pairing
to these target mRNAs. In animals, miRNAs tend to bind to the 3’ untranslated
region (3’ UTR) of their target transcripts to repress translation.
The pairing between miRNAs and their target mRNAs usually includes short
bulges and/or mismatches. In contrast, in all known cases, plant miRNAs
bind to the protein-coding region of their target mRNAs with three or fewer
mismatches and induce target mRNA degradation or repress mRNA translation.
Objectives
- While the first miRNAs were discovered using experimental methods,
experimental miRNA identification remains technically challenging
and incomplete. This calls for the development of computational
approaches to complement experimental approaches to miRNA gene
identification. We propose in this project to investigate
de novo miRNA precursor prediction methods. We follow the
"generation, feature selection, and feature integration"
paradigm of constructing recognition models for genomics sequences.
We generate and identified features based on information in both
primary sequence and secondary structure, and use these features to
construct decision models for the recognition of miRNA precursors.
- Analyzing the binding of microRNAs (miRNA) to their mRNA target
sites reveals that many different factors determine what constitutes
a good fit. We intend to investigate these factors in detail and to
construct decision models for predicting miRNA targets.
- Finally, we would like to understand the role of miRNAs in a
number of human diseases. In particular, we plan to begin our analysis
with genes involved in muscular dystrophy, as this group of genes
are among the largest and most complex-structured human genes.
Selected Publications
- Liang Huai Yang, Wynne Hsu, Mong Li Lee, Limsoon Wong.
SVM-based Identification of microRNA Precursors.
Proceedings of 4th Asia-Pacific Bioinformatics Conference,
pages 267--276, Taipei, Taiwan, February 2006.
PDF
- Yun Zheng, Wynne Hsu, Mong Li Lee, Limsoon Wong.
Exploring Essential Attributes for Detecting MicroRNA Precursors
from Background Sequences.
Proceedings of 2006 VLDB Workshop on Data Mining in Bioinformatics,
pages 131--145, Seoul, Korea, September 2006.
PDF,
Supplementary Info
- Stanley Ng, Santosh Mishra.
Spectral Graph Partitioning Analysis of In Vitro Synthesized
RNA Structural Folding.
Proceedings of International Workshop on Pattern Recognition
in Bioinformatics (PRIB), pages 81--92, Hong Kong, August 2006.
- Stanley Ng, Santosh Mishra.
Unique folding of precursor microRNAs:
Quantitative evidence and implications for de novo identification.
RNA, 13(2):170--187, 2007.
- Stanley Ng, Santosh Mishra.
De novo SVM classification of precursor microRNAs from genomic
pseudo hairpins using global and intrinsic folding measures.
Bioinformatics, 23(11):1321--1330, June 2007.
Dissertations
Acknowledgements
This project is supported in part by the
I2R-SOC Joint Lab on Knowledge Discovery
from Clinical Data (7/03 - 6/07),
NUS OLS Bioinformatics Programme (Ng, Wong: 7/07-12/07),
and an A*STAR AGS scholarship (Ng).
Last updated: 5/4/08, Limsoon Wong.
 Recognition of MicroRNA Precursors and Targets
Recognition of MicroRNA Precursors and Targets Recognition of MicroRNA Precursors and Targets
Recognition of MicroRNA Precursors and Targets