 Recovering missing proteins based on biological complexes
Recovering missing proteins based on biological complexes
Participants:
Wilson Wen Bin Goh,
Weijia Kong,
Hui Peng,
Limsoon Wong
Background
Advancements in proteomics are important to biological and clinical
research because assaying protein identities/quantities paints an
immediate picture of the underlying molecular landscape.
However, proteomics still suffers from incomplete proteome coverage issues
(i.e. not all proteins in a sample are observable in a single screen.)
This gives rise to the "missing-protein problem" (MPP),
which we define as difficulty in observing proteins in a proteome
screen given that they are expected to be present.
Due to MPP, efforts to extend proteome profiling in comparative
or clinical studies, which require consistent protein/peptide
detection (and accurate quantification across an extensive dynamic range),
are rendered less effective.
MPP, as defined here, is different from the missing proteins defined by
the Human Proteome Project (HPP). HPP missing proteins are proteins that
have never been observed in the human proteome but are predicted to be
present in the human proteome due to the presence of a functional gene
sequence (e.g. based on genome assembly or partial transcriptome evidence);
i.e. the HPP notion of missing proteins is not sample specific.
Whereas MPP is focused on proteins that are present in a sample but
not detected in a proteome screen on that sample. That is, MPP is about
filling in "holes" in the proteomic profiling data of patients.
Current imputation methods (for filling holes in proteomic profiling data)
rely on a large set of samples to estimate the correlation between the
abundances of two or more proteins from the entire complement of reported
proteins in these samples. They are inapplicable when there are too
few samples. Moreover, some recent studies on imputation methods have
shown they do not perform well on proteomic profiling data even when
given a non-trivial number of samples.
Objectives
The three main goals of this project are:
- We re-consider the conventional HPP guideline (aka the two-peptide rule)
that a protein is only considered detected when there are at least
two non-bested peptides detected and both uniquely map to that
protein. We investigate whether and how this two-peptide rule can
be relaxed without compromising the reliability of a proteomic screen.
- We propose a novel ranking strategy for missing-protein recovery based
on protein complexes. We postulate that protein complexes provide a
good context for making inference of a protein's presence and its abundance.
This postulate is a significant departure from conventional imputation-based
approaches for filling holes in proteomics profiling data. Notably,
it is applicable for predicting whether a protein is present even when there
is only one sample. This is because the postulate implies on two reasonable
hypotheses, which allow the likelihood of a candidate missing protein
being actually present in a sample to be estimated based on the likelihood
of its parent complexes being present in the sample:
(i) The likelihood of a protein complex being present in a
sample is proportional to the fraction of its constituent proteins that
are reliably reported to be present in that sample; and
(ii) the presence of a protein complex in a sample implies the presence
of all its constituent proteins in that sample. The likelihood of a
protein being present in a sample can then be derived from the likelihoods
of the presence of protein complexes that it is a constituent of.
- We propose that the abundance of a missing protein (which is predicted
to be present in a sample) can be imputed from the abundance of
proteins which are in the same complexes (which are likely
to be present in that sample) as the missing proteins,
as the correlation in the abundance of these proteins with
the missing protein is more likely genuine.
Achievements
- ProInfer (Protein Inference) for
retrieving proteins supported only by ambiguous peptides and
for enhancing the confidences associated with proteins supported by
both unique and ambiguous peptides.
ProInfer’s capabilities was further expanded via integration with
protein complex networks (ProInfer_cpx) to rescue proteins with
weak support from proteomics data. ProInfer achieved excellent
performance especially when a loose filtering is applied on peptides.
In addition, ProInfer runs much faster and requires smaller volumes
of memory but achieves good proteome coverage compared to popular
protein inference tools (Fido, EPIFANY and PIA).
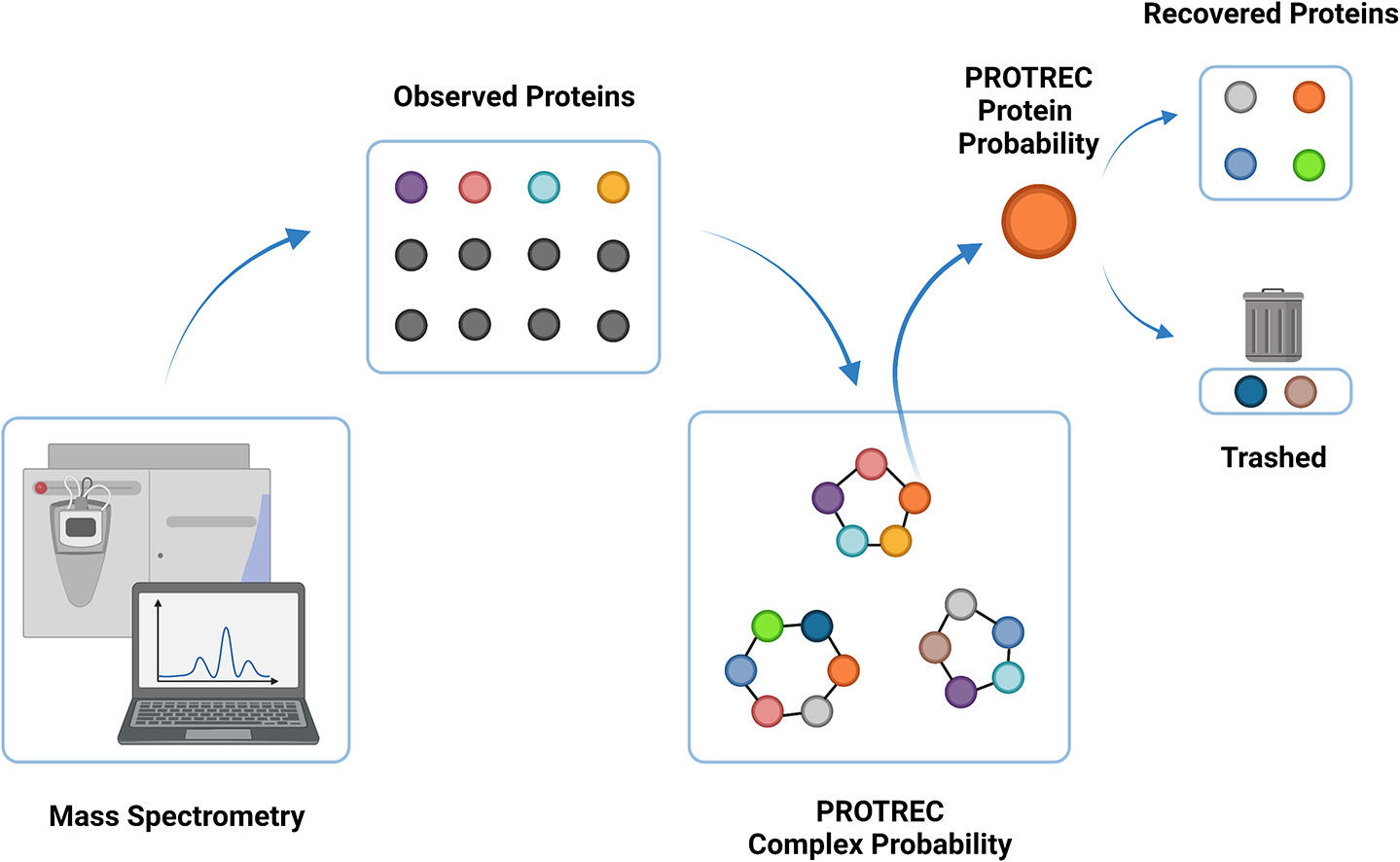
- PROTREC (Protein Recovery) for
predicting missing proteins in a proteomic screen.
The probability of the existence of a complex is estimated
based on the fraction of its proteins reported in the screen.
The probability of an unreported protein in the complex
being actually present are then Bayesian-inferred.
Proteins reported at PROTREC-estimated probability of >90%
are highly verifiable (>90%).
Notably, PROTREC is able to make reliable missing protein calls
given only a single sample.
- ProJect, a novel mixed-model method for missing
value imputation (MVI) for all three kinds of missing values,
viz. MNAR, MCAR, and MAR. ProJect dominated other MVI methods
across a variety of high-throughput data of various modalities
including genomics and mass-spectrometry (MS)-based proteomics.
Unlike other MVI methods that manifest occasional but spectacular errors,
ProJect is very stable.
Selected Publications
- Wilson Wen Bin Goh, Limsoon Wong.
Integrating networks and proteomics: Moving forward.
Trends in Biotechnology, 34(12):951--959, December 2016.
PDF
- Longjian Zhou, Limsoon Wong, Wilson Wen Bin Goh.
Understanding missing proteins: A functional perspective.
Drug Discovery Today, 23(3):644-651, March 2018.
- Weijia Kong, Bertrand Jernhan Wong, Huanhuan Gao, Tiannan Guo,
Xianming Liu, Xiaoxian Du, Limsoon Wong, Wilson Wen Bin Goh.
PROTREC: A probability-based approach for recovering missing
proteins based on biological networks.
Journal of Proteomics, 250:104392, January 2022.
PDF,
PROTREC Server
- Zelu Huang, Weijia Kong, Bertrand Jernhan Wong, Huanhuan Gao,
Tiannan Guo, Xianming Liu, Xiaoxian Du, Limsoon Wong, Wilson Wen Bin Goh.
Proteomic datasets of HeLa and SiHa cell lines acquired by DDA-PASEF
and diaPASEF.
Data in Brief, 41:107919, April 2022.
PDF
- Bertrand Jernhan Wong, Weijia Kong, Limsoon Wong, Wilson Wen Bin Goh.
Resolving missing protein problems using functional class scoring.
Scientific Reports, 12(1):11358, July 2022.
PDF
- Wilson Wen Bin Goh, Weijia Kong, Limsoon Wong.
Evaluating network-based missing protein prediction using p-values,
Bayes factors, and probabilities.
Journal of Bioinformatics and Computational Biology,
21(1):2350005, March 2023.
PDF
- Hui Peng, Limsoon Wong, Wilson Wen Bin Goh.
ProInfer: An interpretable protein inference tool leveraging
on biological networks.
PLoS Computational Biology, 19(3):e1010961, March 2023.
PDF,
ProInfer in Scala
- Wilson Wen Bin Goh, Harvard Wai Hann Hui, Limsoon Wong.
How missing value imputation is confounded with batch effects and
what you can do about it.
Drug Discovery Today, 28(9):103661, September 2023.
PDF
- Weijia Kong, Bertrand Jern Han Wong, Harvard Wai Hann Hui, Kai Peng Lim,
Yulan Wang, Limsoon Wong, Wilson Wen Bin Goh.
ProJect: A powerful mixed-model missing value imputation method.
Briefings in Bioinformatics, 24(4):bbad233, July 2023.
PDF
- Fuchu He, Ruedi Aebersold, Mark S. Baker, et al.
π-HuB: The proteomic navigator of the human body.
Nature, 636:322--331, December 2024.
Dissertations
- Kong Weijia, "Missing protein identification and quantification
using networks", PhD thesis, Nanyang Technological University,
Singapore, submitted 2025.
Selected Presentations
- Limsoon Wong.
Improving coverage and consistency of MS-based proteomics.
Invited talk at 12th Korea-Singapore Joint Workshop on Bioinformatics
and Natural Language Processing,
KAIST, Daejeon, Korea, 22-23 September 2016.
PPT
- Limsoon Wong.
Improving coverage and consistency of MS-based proteomics.
Invited keynote at 16th IEEE International Conference on
Bioinformatics and Bioengineering,
Taichung, Taiwan, 31 October - 2 November 2016.
PPT
- Limsoon Wong.
Advancing clinical proteomics via analysis
based on biological complexes.
Keynote at 16th International Conference on Bioinformatics (InCoB),
Shenzhen, China, 20 - 22 September 2017.
PPT
- Limsoon Wong.
Robustness of protein complex-based analysis of proteomics data.
Invited talk at 1st Westlake Symnposium for Proteomic Big Data,
Westlake University, Hangzhou, China, 30 June 2019.
PPT
Acknowledgements
This project was supported in part by MOE Tier-2 grant MOE2019-T2-1-042.
Last updated: 27/12/2024, Limsoon Wong.
 Recovering missing proteins based on biological complexes
Recovering missing proteins based on biological complexes
 Recovering missing proteins based on biological complexes
Recovering missing proteins based on biological complexes