 Enabling More Sophisticated Proteomic Profile Analysis
Enabling More Sophisticated Proteomic Profile Analysis
Participants:
Wilson Wen Bin Goh, Yie Hou Lee, Limsoon Wong
Background
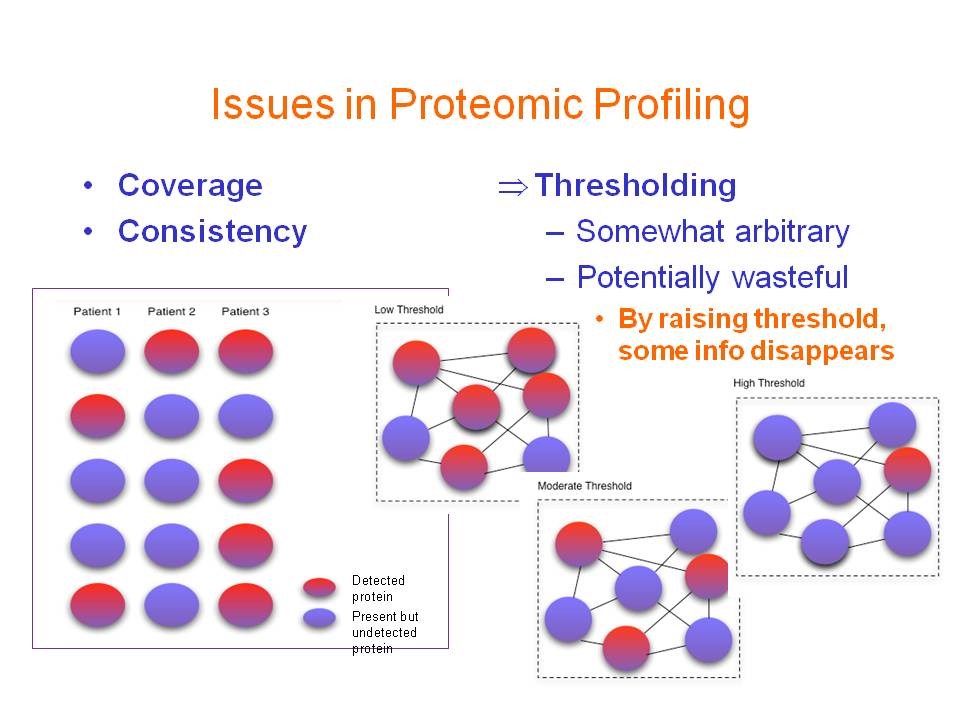
Mass spectrometry (MS)-based proteomics is a powerful tool for profiling
systems-wide protein expression changes. It can be applied for various
purposes, e.g., biomarker discovery in diseases and study of drug responses.
However, MS-based proteomics tend to have consistency (poor reproducibility
and inter-sample agreement) and coverage (inability to detect the entire
proteome) issues that need to be urgently addressed. The former implies
that multiple analytical runs of the same sample under constant
experimental conditions will result in the detection of different but
overlapping sets of proteins. Intuitively, this means more LC-MS/MS runs
are required to identify a sufficiently large portion of any proteome
and is intricately linked to the second issue of inadequate proteome coverage.
Experimental methods to overcome these issues are technically challenging,
resource heavy or place an unreasonable heavy dependency on the quality
of the initial data set. These include exhaustive fractionation of samples,
repeated MS runs of the same sample to reach saturation and compilation of
MS data specific to a sample type generated and archived from different
laboratories. The problems are particularly exemplified in a large-scale
collaborative study to assess the extent of reproducibility across
different laboratories. The results were striking: only 7 out of 27
laboratories correctly reported all 20 proteins, and only 1 laboratory
successfully reported all 22 unique peptides. Therefore, alternative
approaches are needed to complement existing experimental approaches to
circumvent the stochastic sampling of peptides by MS and increase the
comprehensiveness of proteome coverage.
Objectives
In this project, we aim to deal with the two challenges above by proposing
approaches that analyze proteomic profiles in the context of biological
networks. Our two main goals are thus:
- Improving coverage of proteomic profiles.
There are cases where the mass spectra may identify some particular
proteins but, because their scores are below the defined cutoff threshold,
may not be reported initially in the first round of data analysis.
This occurs frequently in the tradeoff between sensitivity and
specificity in precursor ion selection for fragmentation. It is an even
more severe problem in less abundant proteins commonly shrouded in
shotgun proteomics. We propose investigating techniques based on biological
networks to recover such ``undetected'' proteins.
- Improving consistency of proteomic profiles.
Quantitative comparison of samples is central to proteomics. However,
biomarkers identified in one batch are quite often not consistent and not
reproducible in another batch of samples. This is likely due to (i) the
noise and coverage of the proteome at the level of individual samples and
(ii) limitation of current statistical techniques as a result of
insufficient sample size. An analogous situation exists in gene expression
profile analysis, although it is much more severe here---typical proteomic
profiling datasets contain much fewer patients and there are many more
holes (i.e., missing proteins) in the proteomic profile of each patient.
We propose investigating techniques based on biological networks to more
reproducibly and consistently identify biomarkers and achievement more
reliable proteomic-based diagnosis.
In addition, the following two secondary goals are complementary to
and support the two main goals:
- Collecting and integrating data sources of biological pathways,
protein interaction networks, and protein complexes.
The methods that we plan to investigate and develop above rely on the
availability of biological network information in a properly organized
form. However, while there is now a proliferation of pathway databases,
the effectiveness of these databases is hindered by issues such as
incompatible data formats, inconsistent molecular representations,
inconsistent molecular relationship representations, inconsistent
referrals to pathway names, and incomplete data from different
databases.
- Applying the methods developed above to some real-life proteomic
profiling datasets.
At the end of this project, we hope to collaborate with some clinicians
and biologists to apply the methods developed here to new data.
Selected Publications
- Wilson Wen Bin Goh, Yie Hou Lee, Maxey Chung, Limsoon Wong.
How advancement in biological network analysis methods
empowers proteomics.
Proteomics, 12(4--5):550--563, February 2012.
PDF
- Wilson Wen Bin Goh, Yie Hou Lee, Ramdzan Zubaidah, Jingjing Jin,
Difeng Dong, Qingsong Lin, Maxey Chung, Limsoon Wong.
A Network-based pipeline for analyzing MS data---An application towards
liver cancer.
Journal of Proteome Research, 10(5):2261--2272, May 2011.
- Wilson Wen Bin Goh, Yie Hou Lee, Zubaidah Ramdzan, Marek Sergot,
Maxey Chung, Limsoon Wong.
Proteomics Signature Profiling (PSP): A novel contextualization approach
for cancer proteomics.
Journal of Proteome Research, 11(3):1571--1581, March 2012.
PDF
- Wilson Wen Bin Goh, Yie Hou Lee, Zubaidah M. Ramdzan,
Maxey Chung, Limsoon Wong, Marek Sergot.
A network-based maximum link approach towards MS identifies potentially
important roles for undetected ARRB1/2 and ACTB in liver cancer progression.
International Journal of Bioinformatics Research and Applications,
8(3/4):155--170, August 2012.
- Wilson Wen Bin Goh, Mengyuan Fan, Hong Sang Low, Marek Sergot, Limsoon Wong.
Enhancing the utility of Proteomics Signature Profiling (PSP) with
Pathway Derived Subnets (PDSs), performance analysis and
specialised ontologies.
BMC Genomics, 14:35, February 2013.
PDF
- Wilson Wen Bin Goh, Marek Sergot, Judy Sng, Limsoon Wong.
Comparative network-based recovery analysis and proteomic profiling of
neurological changes in valporic acid-treated mice.
Journal of Proteome Research, 12(5):2116--2127, April 2013.
PDF
- Yie Hou Lee, Wilson Wen Bin Goh, Choon Keow Ng, Manfred Raida, Limsoon Wong,
Qingsong Lin, Urs A. Boelsterli, Maxey Chung.
Integrative toxicoproteomics implicates impaired mitochondrial
glutathione import as off-target effect of troglitazone.
Journal of Proteome Research, 12(6):2933--2945, May 2013.
- Wilson Wen Bin Goh, Limsoon Wong.
Networks in proteomic analysis of cancer.
Current Opinion in Biotechnology, 24(6):1122--1128, December 2013.
PDF
- Wilson Wen Bin Goh, Limsoon Wong, Judy Chia Ghee Sng.
Contemporary network proteomics and its requirements.
Biology, 3(1):22--38, December 2013.
PDF
- Wilson Wen Bin Goh, Limsoon Wong.
Computational proteomics: Designing a comprehensive analytical strategy.
Drug Discovery Today, 19(3):266-274, March 2014.
-
Mengyuan Fan, Hong Sang Low, Hufeng Zhou, Markus R. Wenk, Limsoon Wong.
LipidGO: Database for lipid-related GO terms and applications.
Bioinformatics, 30(7):1043--1044, April 2014.
PDF
- Mengyuan Fan, Hong Sang Low, Markus Wenk, Limsoon Wong.
A semi-automated methodology for finding lipid-related GO terms.
Database, 2014:bau089, September 2014.
PDF
- Hirotaka Oikawa, Wilson Wen Bin Goh, Vania Lim, Limsoon Wong, Judy Sng.
Valporic acid mediates BDNF through miR-124 by down-regulating a
novel protein target, GNAI1.
Neurochemistry International, 91:62--71, December 2015.
PDF
- Wilson Wen Bin Goh, Tiannan Guo, Ruedi Aebersold, Limsoon Wong.
Quantitative proteomics signature profiling
based on network contextualization.
Biology Direct, 10:71. December 2015.
PDF
- Wilson Wen Bin Goh, Limsoon Wong.
Design principles for clinical network-based proteomics.
Drug Discovery Today, 21(7):1130--1138, July 2016.
- Wilson Wen Bin Goh, Limsoon Wong.
Advancing clinical proteomics via analysis based on biological
complexes: A tale of five paradigms.
Journal of Proteome Research, 15(9):3167--3179, July 2016.
NetProt v0.1
- Wilson Wen Bin Goh, Limsoon Wong.
Evaluating feature-selection stability in next-generation proteomics.
Journal of Bioinformatics and Computational Biology,
14(5):1650029, October 2016.
PDF
- Wilson Wen Bin Goh, Limsoon Wong.
Spectra-first feature analysis in clinical proteomics---A case
study in renal cancer.
Journal of Bioinformatics and Computational Biology,
14(5):1644004, October 2016.
PDF
- Wilson Wen Bin Goh, Limsoon Wong.
Integrating networks and proteomics: Moving forward.
Trends in Biotechnology, 34(12):951--959, December 2016.
PDF
- Wilson Wen Bin Goh, Limsoon Wong.
Protein complex-based analysis is resistant to the obfuscating
consequences of batch effects---a case study in clinical proteomis.
BMC Genomics, 18(Suppl 2):142, March 2017.
PDF
- Wilson Wen Bin Goh, Limsoon Wong.
Class-paired Fuzzy SubNETs: A paired variant of the rank-based network
analysis family for feature selected based on protein complexes.
Proteomics, 17(10):1700093, May 2017.
- Wilson Wen Bin Goh, Limsoon Wong.
NetProt: Complex-based feature selection.
Journal of Proteome Research, 16(8):3102--3112, June 2017.
NetProt v0.1
- Longjian Zhou, Limsoon Wong, Wilson Wen Bin Goh.
Understanding missing proteins: A functional perspective.
Drug Discovery Today, 23(3):644-651, March 2018.
- Wilson Wen Bin Goh, Limsoon Wong.
Advanced bioinformatics methods for practical applications in proteomics.
Briefings in Bioinformatics, 20(1):347--355, January 2019.
PDF
- Wilson Wen Bin Goh, Yaxing Zhao, Andrew Chi-Hau Sue,
Tiannan Guo, Limsoon Wong.
Proteomic investigation of intra-tumor heterogeneity using network-based
contextualization---a case study on prostate cancer.
Journal of Proteomics, 206:103446, August 2019.
PDF
Dissertations
Selected Presentations
- Limsoon Wong.
Analysis of Gene Expression and Proteomic Profiles
based on Biological Networks.
Tutorial at 10th Asia Pacific Bioinformatics Conference (APBC2012),
Melbourne, Australia, 17-19 January 2012.
PPT,
Written notes
- Limsoon Wong.
Using Biological Networks for Protein Function Prediction,
Biomarker Identification, and Other Problems in Computational Biology.
Invited master class at 2012 International Winter School in Methods
in Bioinformatics (WSMBio 2012),
Tarragona, Spain, 20-24 February 2012.
PPT
- Limsoon Wong.
A Novel Contextualization Approach to Proteomic Profile Analysis.
Invited talk at Global-COE Workshop on Engineering/Information Science
for Integrated Life Science and Predictive Medicine,
Hotel Grand Park City Hall, Singapore, 28 February 2012.
PPT
- Limsoon Wong.
Living with Noise.
Invited talk at SinFra 2012,
UPMC, 15 October 2012.
PPT
- Limsoon Wong.
A novel contextualization approach to proteomic profile analysis.
Invited plenary talk at 2nd Biomarker Discovery Conference (BDC2012),
Shoal Bay, Australia, 6 December 2012.
PPT
- Limsoon Wong.
Improving Proteomic Profile Analysis by Contextualization.
Invited talk at 3rd IPM-NUS Workshop on Computational Biology,
Institute for Research in Fundamental Sciences (IPM), Tehran, Iran,
25 February 2013.
PPT
- Limsoon Wong.
The use of context in gene expression and proteomic profile analysis.
Invited talk at
NUS-ENS Workshop on Novel Genome-Wide Approaches to Decipher Transcriptional
and Epigenetic Regulation in Mammalian Cells,
Paris, France, 30-31 May 2013.
PPT
- Limsoon Wong.
The use of context in gene expression and proteomic profile analysis.
Invited master class at
2013 International Summer School on Trends in Computing (SSTiC2013),
Tarragona, Spain, 22-26 July 2013.
PPT
- Limsoon Wong.
Disease gene expression and proteomic profile analysis based on regulatory
networks and systems.
Invited master class at
EMS Autumn School on Computational Aspects of Gene Regulation,
Bedlewo, Poland, 13-19 October 2013.
PDF
- Limsoon Wong.
Computational thinking in genome and proteome analysis.
Invited talk at International Conference on Next Generation
Genomic View on Plants, Animals, and Microbes,
National University of Singapore, 5 March 2014.
PPT
- Limsoon Wong.
Advancing clinical proteomics using protein complexes as a
contextualization framework.
Invited talk at Networks in Biological Sciences:
The Protein Network Workshop,
Institute for Mathematical Sciences, Singapore, 8 June 2015.
PPT
- Limsoon Wong.
Network-based analysis of proteomics profiles.
Keynote talk at KAUST Research Conference on Computational
and Experimental Interfaces of Big Data and Biotechnology,
Thuwal, Saudi Arabia, 25-27 January 2016.
PPT
- Limsoon Wong.
Enabling more sophisticated proteomic profile analysis.
Invited talk at the St. Petersburg International Symposium on
Systems Biology and Bioinformatics (SBBI'2016),
St. Petersburg, 30 June - 2 July 2016.
PPT
- Limsoon Wong.
Enabling more sophisticated analysis of proteomics profiles.
Invited talk at the Workshop on Gene Dynamics and Chromosomes,
IAS, City University of Hong Kong, 11-15 August 2016.
PPT
- Limsoon Wong.
Improving coverage and consistency of MS-based proteomics.
Invited talk at 12th Korea-Singapore Joint Workshop on Bioinformatics
and Natural Language Processing,
KAIST, Daejeon, Korea, 22-23 September 2016.
PPT
- Limsoon Wong.
Improving coverage and consistency of MS-based proteomics.
Invited keynote at 16th IEEE International Conference on
Bioinformatics and Bioengineering,
Taichung, Taiwan, 31 October - 2 November 2016.
PPT
- Limsoon Wong.
Advancing clinical proteomics via analysis
based on biological complexes.
Keynote at 16th International Conference on Bioinformatics (InCoB),
Shenzhen, China, 20 - 22 September 2017.
PPT
- Limsoon Wong.
Robustness of protein complex-based analysis of proteomics data.
Invited talk at 1st Westlake Symnposium for Proteomic Big Data,
Westlake University, Hangzhou, China, 30 June 2019.
PPT
Acknowledgements
This project is supported in part by
NRF CRP grant NRF-G-CRP-2997-04-082(d),
A*STAR PSF grant SERC 102 101 0030,
MOE Tier-2 grant MOE2012-T2-1-061, and
MOE Tier-2 grant MOE2019-T2-1-042.
Last updated: 29/8/2019, Limsoon Wong.
 Enabling More Sophisticated Proteomic Profile Analysis
Enabling More Sophisticated Proteomic Profile Analysis
 Enabling More Sophisticated Proteomic Profile Analysis
Enabling More Sophisticated Proteomic Profile Analysis